Full Workflow: Estimate Gene Expression Entropy
Aryan Kamal, Charles Dussiau, Judith Zaugg
25 April 2025
workflow.RmdThis vignette demonstrates a complete example of how to use the
Entropy package to compute entropy values for cells in a
Seurat object.
📦 Load libraries and data
## Loading required package: SeuratObject## Loading required package: sp##
## Attaching package: 'SeuratObject'## The following objects are masked from 'package:base':
##
## intersect, t
data("pbmc_small")
DefaultAssay(pbmc_small) <- "RNA"
result <- run_entropy(
seu = pbmc_small,
assay = "RNA",
nn_list = NULL, # Automatically computes neighbors
output_path = NULL, # Don’t save the full matrix for now
add_assay = TRUE # Add entropy matrix as a new assay
)## INFO [2025-04-25 14:45:15] Starting entropy estimation for assay: RNA
## INFO [2025-04-25 14:45:15] Input Seurat object contains 80 cells and 230 features
## INFO [2025-04-25 14:45:15] Assay 'RNA' found and set as default assay.
## INFO [2025-04-25 14:45:15] No neighbor list provided. Computing neighbors using PCA reduction.## Computing nearest neighbors## Only one graph name supplied, storing nearest-neighbor graph only## INFO [2025-04-25 14:45:16] Selecting top 3000 variable features.## Warning: The `slot` argument of `GetAssayData()` is deprecated as of SeuratObject 5.0.0.
## ℹ Please use the `layer` argument instead.
## ℹ The deprecated feature was likely used in the Seurat package.
## Please report the issue at <https://github.com/satijalab/seurat/issues>.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.## INFO [2025-04-25 14:45:20] Entropy matrix added to Seurat object as 'RNA_entropy'.
# Check the added assay
result[["RNA_entropy"]]## Assay data with 230 features for 80 cells
## First 10 features:
## MS4A1, CD79B, CD79A, HLA-DRA, TCL1A, HLA-DQB1, HVCN1, HLA-DMB, LTB,
## LINC00926
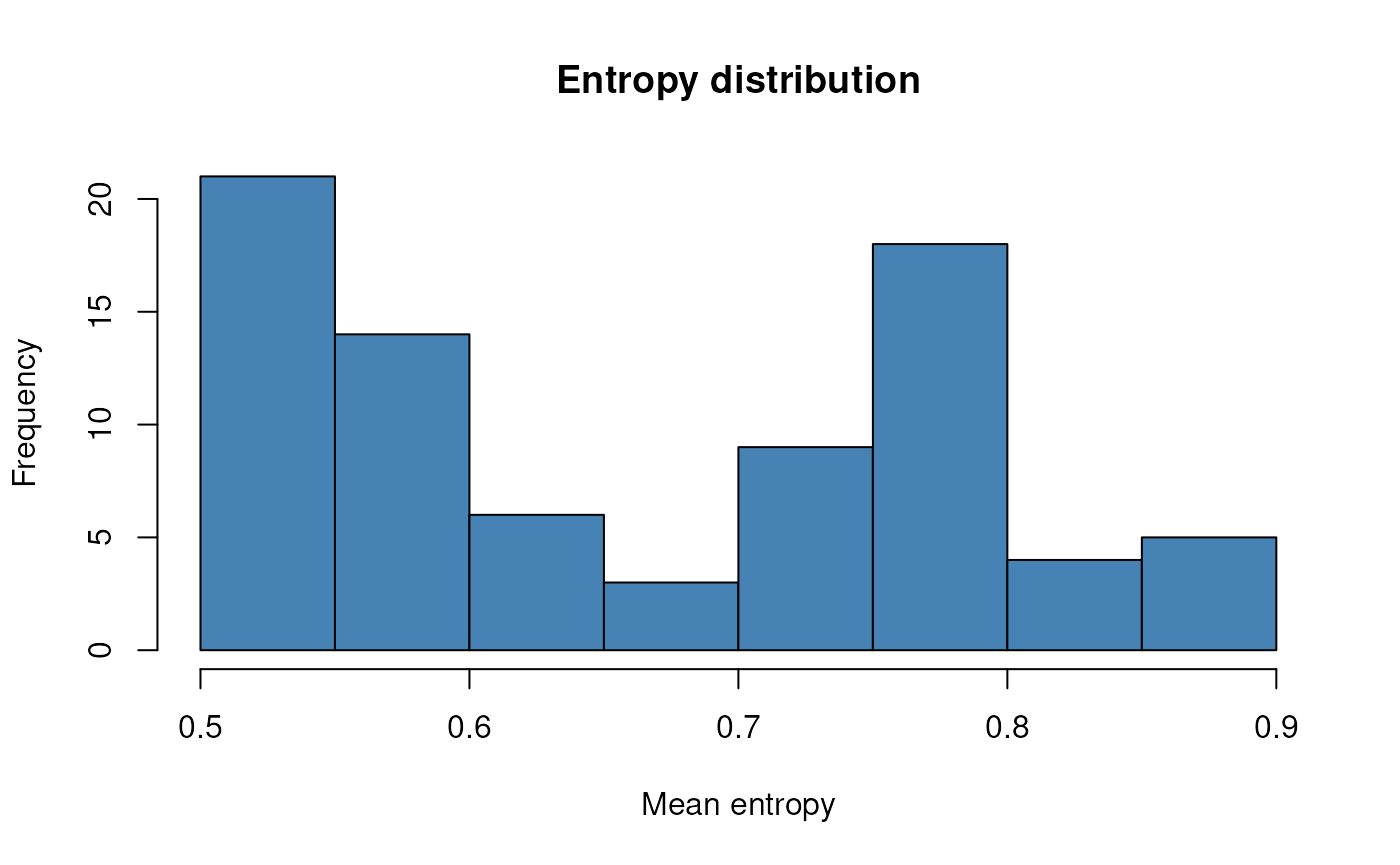
# Visualize mean entropy per cell
means <- colMeans(GetAssayData(result[["RNA_entropy"]]))
hist(means, main = "Entropy distribution", xlab = "Mean entropy", col = "steelblue")